Jupyter Notebook¶
interactive computing
cells can be a code, markdown or raw text
prints out last time of cell, no need to add
print()use markdown to write your thoughts
two modes: command mode and edit mode
shortcuts to remember:¶
enterordouble-click, start edit modeescto return to command modeshift+enterexecute current cell and move to nextctrl+enterexecute current cell and stay thereaandb, add cell above or belowdddelete a cellcand thenv, copy and paste a cellmturn cell into markdownytrun cell into code
The basics¶
# no data type declaration
a = 3
b = 19
a+b
22
# string concat
first_name = 'Shaji'
second_name = 'P'
first_name+' '+second_name
'Shaji P'
# string method example
statement = "this is a sentence"
statement.count('s')
3
# string method example
statement.split()
['this', 'is', 'a', 'sentence']
# Python list
x = [12,9,6,4]
y = [1,2,4]
x+y # lists are concatenated
[12, 9, 6, 4, 1, 2, 4]
# list method example
z = x+y
z.count(4)
2
# in-build sum function
sum(x)
31
# in-built sort
sorted(x)
[4, 6, 9, 12]
# using functions from math
from math import pi,sqrt
r = 4
sqrt(2*pi*r)
5.0132565492620005
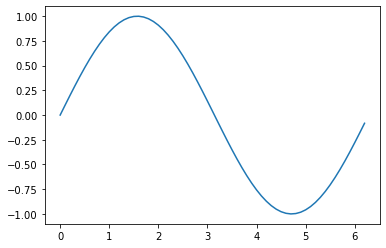
Plotting¶
import matplotlib.pyplot as plt
import numpy as np # see next section
%matplotlib inline
# create range of values from 0 to 2pi in steps of 0.1
x = np.arange(0,2*pi,0.1)
# create y as sine function with x as independent variable
y = np.sin(x)
# graph for the function y
plt.plot(x,y)
[<matplotlib.lines.Line2D at 0x7f1d0d5e3f70>]
Numpy¶
# numpy, the backborne of scientific computing
# all array related operations are defined in numpy
import numpy as np
# create 2x3 array of 1's
x_arr = np.ones((2,3))
x_arr
array([[1., 1., 1.],
[1., 1., 1.]])
# adds a scalar value element wise
x_arr + 4
array([[5., 5., 5.],
[5., 5., 5.]])
x_arr
# to reflect change, store the values to the old array
# uncomment below two lines to see the change
#x_arr = x_arr+4
#x_arr
array([[1., 1., 1.],
[1., 1., 1.]])
y_arr = np.array([2,2,2])
y_arr
array([2, 2, 2])
# array broadcasting
# matches the shape and adds the y_arr row to each -
# row of x_arr
x_arr + y_arr
array([[3., 3., 3.],
[3., 3., 3.]])
y_arr = np.array([5,8])
x_arr+y_arr # broadcasting fails
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
<ipython-input-20-57fdba28a423> in <module>
1 y_arr = np.array([5,8])
----> 2 x_arr+y_arr # broadcasting fails
ValueError: operands could not be broadcast together with shapes (2,3) (2,)
# to rectifyabove error and add elements of y_arr to each column of x_arr
# change "orentation of y_arr" first
y_arr[:,np.newaxis]
array([[5],
[8]])
# now you can add them
y_arr[:,np.newaxis] + x_arr
array([[6., 6., 6.],
[9., 9., 9.]])
Speeding up operations with code change¶
import random
import numba
# create a list of 10k random elements
x = [random.random() for i in range(10000)]
y = [random.random() for i in range(10000)]
z = [] # empty list to store result
%%time
# first, let's try good old for loop
for i in range(len(x)):
z.append(x[i] + y[i])
print(z[:3]) # print first 3 elements
[1.1532859016438493, 1.334577514000911, 0.3511312253827581]
CPU times: user 1.14 ms, sys: 416 µs, total: 1.56 ms
Wall time: 1.56 ms
%%time
# now list comprehension
z = [x[i] + y[i] for i in range(len(x))]
CPU times: user 927 µs, sys: 0 ns, total: 927 µs
Wall time: 931 µs
%%time
# using zip()
# zip() and enumerate() are useful functions
z = [a + b for a,b in zip(x,y)]
CPU times: user 747 µs, sys: 271 µs, total: 1.02 ms
Wall time: 1.02 ms
# create numpy arrays
xa = np.array(x)
ya = np.array(y)
%%time
# using numpy addition
za = xa+ya
za[:3]
CPU times: user 67 µs, sys: 25 µs, total: 92 µs
Wall time: 94.7 µs
array([1.1532859 , 1.33457751, 0.35113123])
# Take another example of finding sum of all elements in an array
# Below function finds sum of all elements in x
def add(x):
total = 0
for i in range(x.shape[0]):
total = total+x[i]
return total
# array of 10 million items
x = np.random.rand(10000000)
%%time
add(x)
CPU times: user 2.04 s, sys: 0 ns, total: 2.04 s
Wall time: 2.04 s
4998375.354010175
Just in time (JIT) compiler¶
@numba.jit
def add_jit(x):
total = 0
for i in range(x.shape[0]):
total = total+x[i]
return total
%%time
add_jit(x)
CPU times: user 175 ms, sys: 524 µs, total: 176 ms
Wall time: 175 ms
4998375.354010175
%%time
add_jit(x) # already compiled, hence faster this time
CPU times: user 11.9 ms, sys: 214 µs, total: 12.1 ms
Wall time: 12.2 ms
4998375.354010175
%%time
# numpy sum
x.sum()
CPU times: user 5.6 ms, sys: 0 ns, total: 5.6 ms
Wall time: 5.04 ms
4998375.354010154
Remarks:¶
Python is not slow per say
the way you code matters
stick to existing fuctions in numpy when available
numpy functions are optimized for speed